The Pathway Tools API
October 21, 2009
Contents
1 Introduction
1.1 Conventions and Notation
2 Software Architecture of Pathway Tools
2.1 Common Lisp
2.2 Franz’s Allegro Common Lisp
2.3 Ocelot Frame Representation System
2.4 Generic Frame Protocol
2.4.1 Terminology
2.5 Pathway Tools Application Code
2.5.1 Pathway Tools API Functions
5 API Functions
5.1 Enumeration Operations
5.1.1 Operations on Reactions
5.1.2 Operations on Pathways
5.1.3 Operations on Enzymatic-Reactions
5.1.4 Operations on Proteins
5.1.5 Operations on Genes
5.1.6 Operations on Regulation Frames
5.1.7 Operations on Compounds
5.1.8 Object Name Manipulation Operations
6 Code Examples
6.1 Example Table-Generating Functions
6.2 Example Queries
1 Introduction
The Pathway Tools software suite is more than just a way to view a multidimensional genome annotation, or a way to build Pathway/Genome Databaes (PGDBs). It is also a robust platform for bioinformatics programming. The aim of this document is to explain the Lisp API for Pathway Tools to end-users. Pathway Tools has hundreds of thousands of lines of code, developed over 15 years. There are dozens of high-level computations, and many utility functions, that are ready for use and save countless hours of programming time if used correctly. Those who learn how to programmatically access the Lisp API can unlock the full potential and benefit from using Pathway Tools.
1.1 Conventions and Notation
If we are referring to a Common Lisp symbol, code, or object, we will
often give it a special highlighting. For example, to represent the
frame name of the class that represents reactions, we use the notation
|Reactions|. This is how one would specify the frame name of
the class when writing Common Lisp code for use with the Pathway Tools
API.
Common Lisp code is also given a special highlighting. A short example
would be (all‑rxns :all). Please see the
section on example code to see larger examples of how this would appear.
In the description of arguments to functions, an Optional
argument without a described default value should be assumed to be 'nil
by default. Also, if a function is described as having an Optional
argument, that means that the logic of the function does not require
that the argument be set. It does not necessarily mean a reference to
a Common Lisp &optional argument. On the other hand,
if a function’s argument is described as being a Keyword
argument, it is explicitly in reference to it being a Common
Lisp &keyword argument. Such arguments must be supplied
after the Keyword argument name, where the argument name has
been prefixed with a colon, ‘:’, such as the keyword
allow‑modified‑forms? in the function
all‑transcription‑factors, which would take the form of the
following example function call: (all‑transcription‑factors
:allow‑modified‑forms? nil).
Common Lisp also has a notation for describing symbol names that
contain alphabetic characters that are not upper-case. For example, a
class might have the name of “Protein-Complexes”. The frame ID for
that class would be described as |Protein‑Complexes|. The
vertical bars on either side of the frame ID tells Common Lisp that
the symbol name has lower-case letters, and they shouldn’t be
converted to their upper-case equivalents. This implies that Common
Lisp is not case-sensitive when it comes to frame IDs. For example,
Protein‑Complexes would not be considered the same as
|Protein‑Complexes|, since the former notation would be
indistinguishable from PROTEIN‑COMPLEXES from the perspective
of Common Lisp.
2 Software Architecture of Pathway Tools
Pathway Tools has a layered design. Depending on the data manipulation task at hand, you might want or need to use functions that reside at various levels throughout the architecture. The following sections describe the individual layers.
2.1 Common Lisp
At the most fundamental level, Pathway Tools is built using a
programming language called Common Lisp (CL). CL is a very high-level
robust industrial-strength programming language, with a powerful
object system and integrated compiler. It easily holds its own against
other computer languages, such as C++ and Java, and in
fact is often a
superior choice.
In order to understand the Lisp API of Pathway Tools, you will
first need to understand a bit about Common Lisp. The definitive
source of information on Common Lisp is the ANSI standard. It is
conveniently available online in an extensively hyperlinked document
known as the
Common
Lisp HyperSpec. A more readable and thorough explanation of Common
Lisp can be found in Guy Steele’s "Common Lisp the Language", which is
now
available
online.
Two recently published books on Common Lisp programming
include Paul Graham’s ANSI
Common Lisp, and Peter Seibel’s “Practical Common Lisp”. We
recommend Pathway Tools users that are new to read the latter book,
since it is full of easy to follow exercises, and is available in its
entirety online, for free.
Almost all functions defined in the ANSI Common Lisp standard are accessible from the Pathway Tools Lisp API.
2.2 Franz’s Allegro Common Lisp
The particular Common Lisp implementation that is used in building Pathway Tools is that from Franz. Almost all functions that are available from Allegro Common Lisp (see the Allegro Common Lisp documentation for a list of functions) are also exposed in the Pathway Tools Lisp API. Certain libraries available in the professional version of Allegro Common Lisp are not built in to the Pathway Tools runtime images, due to licensing restrictions.
2.3 Ocelot Frame Representation System
Ocelot is a frame representation system implemented on top of Common Lisp. It can be thought of as an object-oriented database. The term for a collection of classes and instances defined in a frame representation system is a “knowledgebase”. The Ocelot Manual defines a set of functions that are exposed in the Pathway Tools Lisp API. Please see the following for more discussion on frame representation systems The Design Space of Frame Knowledge Representation Systems.
2.4 Generic Frame Protocol
The Generic Frame Protocol (GFP) is a standard API for interacting with frame representation systems. It performs an analogous role as the SQL language performs for different relational databases. The Pathway Tools Lisp API uses GFP operations to interact with the Ocelot frame representation system (though there are a few functions defined in the Ocelot Manual that do not have analogs in GFP). We recommend that those who want to learn how to interact with Pathway Tools read “Introduction”, “A Reference Model of Frame Representation Primitives” (ignore “FRS Behaviors”), “The Generic Frame Protocol” (through “Facet Functions”) of the GFP Specification.
A useful tool for browsing knowledgebases that support GFP would be the Generic Knowledge-Base (GKB) Editor, which is built-in to Pathway Tools.
2.4.1 Terminology
There are certain terms that are used when describing objects, meta-objects, and relationships in a frame system. Much of the terminology is similar to the concepts from object oriented design. Here we provide definitions for the most common occurrences.
- knowledgebase
- A collection of class and instance objects that describe a domain.
- class
- An abstract form of an object. These are arranged in a hierarchy or directed acyclic graph (i.e., multiple inheritance).
- subclass
- A class that is subsumed by one or more other classes.
- superclass
- A class that subsumes one or more other classes.
- instance
- A concrete frame, representing a specific object.
- parent
- The direct or indirect classes that the given frame inherits from. This can follow a transitive closure all the way to the root classes.
- child
- The direct or indirect classes that the given class subsumes. This can follow a transitive closure all the way to the instances.
- slot
- An attribute or member of the frame. These are defined in
their own frames, known as
slot‑units. - facet
- A speficic attribute of the slot.
- annotation
- Data that is associated with a particular value in a particular slot.
- slot-unit
- A frame that defines the properties of a slot, as the slot appears in other frames.
2.5 Pathway Tools Application Code
2.5.1 Pathway Tools API Functions
In the application code level, there are certain functions that have been identified as being useful for end-users as well. These functions are exposed in the Pathway Tools Lisp API, and they are described in this document in Section 5.
3 The Pathway Tools Schema
The Pathway Tools schema is the particular collection of classes, and the defined relationships between them, that are present in the knowledgebase of any PGDB. Ocelot supports multiple inheritance of classes, so a class or instance might have more than one direct superclass. An easy way to visualize the Pathway Tools schema is by means of the GKB Editor.
The Pathway Tools schema can be extended by means of GFP operations, but one must be aware that such changes might be destroyed when a PGDB schema upgrade occurs. It is highly recommended to perform any desired addition of classes via the Pathway Tools Navigator if possible, or through the GKB Editor.
Another important concept is that of a slot‑unit. For
any given slot in the schema, there is a special frame known as a
slot‑unit, with the name of the slot as the frame ID. The
slot‑unit frame defines the properties of the slot, known as
facets. Please see the Ocelot and GFP documentation for more
information.
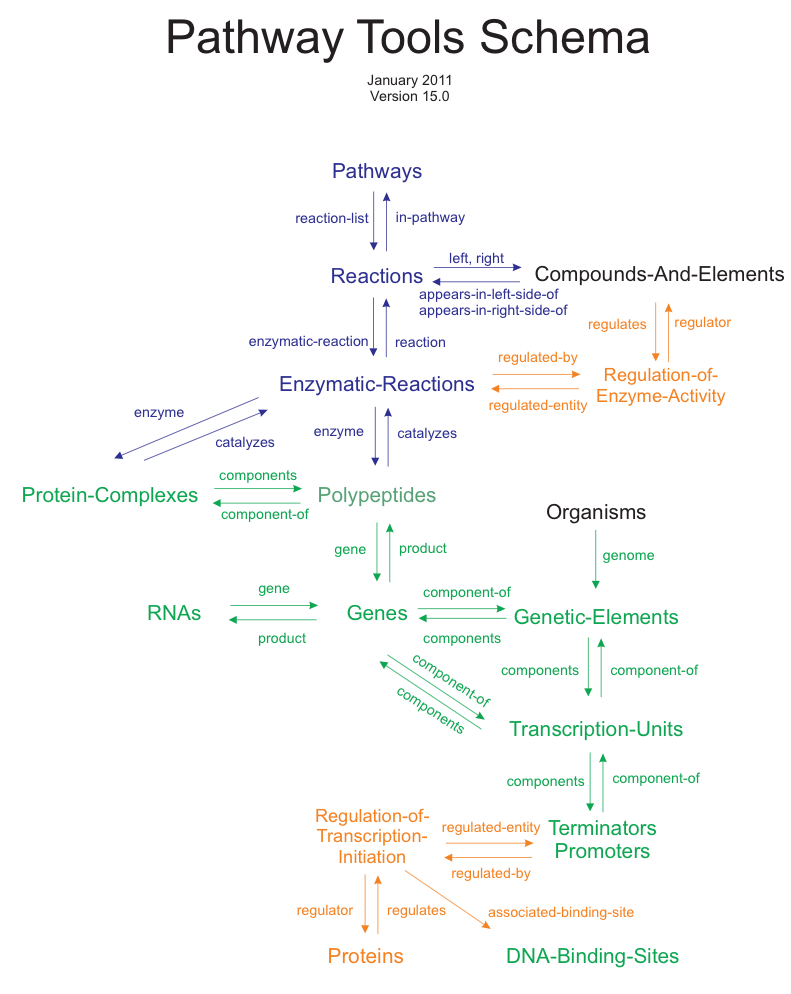 |
| Figure 1: Major relationships among the major classes of the Pathway Tools schema. Colors indicate biological areas: blue for reaction and pathway information, green for genome and protein information; orange for regulation. |
4 Getting Started
4.1 Caveats
One aspect of GFP is that it defines a notion of the “current knowledgebase”. This is similar to how many relational database management systems work, in that any query must be against a specified database/schema, and queries default to the “current” database/schema. In the context of Pathway Tools, every PGDB is a organism-specific knowledgebase. For many functions, it assumes that the operations are to be performed on the “current organism” (i.e., the selected PGDB). Some functions allow you to provide an optional argument, where you can specify which organism you wish to have the function run against.
5 API Functions
This section lists a series of functions that are used for convenient navigation from one object type to another, such as from biochemical pathways to their constituent reactions, or from DNA binding sites to regulation description objects.
Unless otherwise specified, it should be assumed that all of these functions apply only to the currently selected PGDB.
5.1 Enumeration Operations
- all-pathways
-
- Description
- Returns a list of pathway instance frames of a specified type.
- Arguments
-
- selector
- Optional Selects whether all pathways, or just small-molecule metabolism base pathways. Can take either a symbol of
:allor:small‑molecule. Defaults to <code>:all</code>. - base?
- Optional If this argument evaluates to true, only includes base pathways. Otherwise, all pathways, including superpathways, will be returned.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Pathways|.
- all-rxns
-
- Description
- Returns a set of reactions that fall within a particular category.
- Arguments
-
- type
- Optional The type of reaction to
return. Defaults to
:metab‑smm. The possible values are:- :all
- All reactions.
- :metab-pathways
- All reactions found within metabolic pathways. Includes reactions that are pathway holes. May include a handfull of reactions whose substrates are macromolecules, e.g., ACP. Excludes transport reactions.
- :metab-smm
- All reactions of small molecule
metabolism, whether or not they are present in a
pathway. Subsumes
:metab‑pathways. - :metab-all
- All enzyme-catalyzed
reactions. Subsumes
:metab‑smm. - :enzyme
- All enzyme-catalyzed reactions (i.e., instances of either EC-Reactions class or Unclassified-Reactions class).
- :transport
- All transport reactions.
- :small-molecule
- All reactions whose substrates are all small molecules, as opposed to macromolecules. Excludes transport reactions.
- :protein-small-molecule-reaction
- One of the substrates of the reaction is a macromolecule, and one of the substrates of the reaction is a small molecule.
- :protein-reaction
- All substrates of the reaction are proteins.
- :trna-reaction
- One of the substrates of the reaction is a tRNA.
- :spontaneous
- Spontaneous reactions.
- :non-spontaneous
- Non-spontaneous reactions that
are likely to be enzyme catalyzed. Some reactions will
be returned for type
:non‑spontaneousthat will not be returned by:enzyme.
- Side-Effects
- None.
- Return Value(s)
- A list of reaction frames.
- all-substrates
-
- Description
- Returns all unique substrates used in the reactions specified
by the argument
rxns. - Arguments
-
- rxns
- A list of reaction frames.
- Side-Effects
- None.
- Return Value(s)
- A list of compound frames. There might be strings in the list,
as the
'leftand'rightslots of a reaction frame can contain strings.
- all-cofactors
-
- Description
- Return a list of all cofactors used in the current PGDB.
- Arguments
- None.
- Side-Effects
- None.
- Return Value(s)
- A list of cofactor frames.
- all-modulators
-
- Description
- Enumerate all of the modulators, or direct regulators, in the current PGDB.
- Arguments
- None.
- Side-Effects
- None.
- Return Value(s)
- A list of regulator frames.
- all-transported-chemicals
-
- Description
- Enumerates all chemicals transported by transport reactions in the current PGDB.
- Arguments
-
- from-compartment
- Keyword, Optional The compartment that the chemical is coming from (see Cellular Component Ontology).
- to-compartment
- Keyword, Optional The compartment that the chemical is going to (see Cellular Component Ontology).
- show-compartment
- Keyword, Optional Show the form of the compound as it is present in the specified compartment (see Cellular Component Ontology), if the compound is modified during transport.
- primary-only?
- Keyword, Optional If true, filter out common transport compounds, such as protons and Na+.
- Side-Effects
- None.
- Return Value(s)
- A list of compound frames.
- all-protein-complexes
-
- Description
- Enumerates different types of protein complexes.
- Arguments
-
- filter
-
Keyword The type of protein complexes to return.
The argument must be one of the following values:
- :all or t
- Return all protein complexes.
- :hetero
- Return all heteromultimers.
- :homo
- Return all homomultimers.
- Side-Effects
- None.
- Return Value(s)
- A list of protein complex frames.
- all-transcription-factors
-
- Description
- Enumerates all transcription factors, or just unmodified forms of transcription factors.
- Arguments
-
- Side-Effects
-
- allow-modified-forms?
- Keyword, Optional A
boolean value. If true, modified and unmodified forms of
the protein are returned. If false, then only unmodified
forms of the proteins are returned. The default value is
t.
- Return Value(s)
- None.
A list of protein frames that are transcription factors.
- all-genetic-regulation-proteins
-
- Description
- Enumerates all proteins that are involved in genetic regulation of a particular given class. Optionally, just unmodified forms of the proteins are returned.
- Arguments
-
- class
- Keyword, Optional
The class
|Regulation|or a subclass. It defaults to|Regulation‑of‑Transcription‑Initiation|. - allow-modified forms?
- Keyword, Optional A
boolean value. If true, modified and unmodified forms of
the protein are returned. If false, then only unmodified
forms of the proteins are returned. The default value is
t.
- Side-Effects
- None.
- Return Value(s)
- A list of protein frames that are involved in the specified form of regulation.
- all-sigma-factors
-
- Description
- Enumerate all RNA polymerase sigma factors.
- Arguments
- None.
- Side-Effects
- None.
- Return Value(s)
- A list of all instances of the class
|Sigma‑Factors|.
- all-operons
-
- Description
- Enumerates all operons. In this case, an operon is defined as
a list of overlapping instances of
|Transcription‑Units|. - Arguments
- None.
- Side-Effects
- None.
- Return Value(s)
- A list of lists of
|Transcription‑Units|, where all|Transcription‑Units|in the list belong to the same operon.
- rxns-w-isozymes
-
- Description
- Enumerate all reactions that have isozymes (distinct proteins or protein classes that catalyze the same reaction).
- Arguments
-
- rxns
- Keyword, Optional A list of instances
of the class
|Reactions|. Defaults to the result of(all‑rxns :enzyme).
- Side-Effects
- None.
- Return Value(s)
- A list of A list of instances
of the class
|Reactions|with isozymes.
- rxns-catalyzed-by-complex
-
- Description
- Enumerates all reactions catalyzed by an enzyme that is a protein complex.
- Arguments
-
- rxns
- Keyword, Optional A list of instances
of the class
|Reactions|. Defaults to the result of(all‑rxns :enzyme).
- Side-Effects
- None.
- Return Value(s)
- A list of instances
of the class
|Reactions|with a protein complex as an enzyme.
- all-enzymes
-
- Description
- Return all enzymes of a given type.
- Arguments
-
- type
- Keyword, Optional
A type as taken from the argument to
#'enzyme?. Defaults to:chemical‑change.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
- all-transporters
-
- Description
- Enumerate all transport proteins.
- Arguments
- None.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
5.1.1 Operations on Reactions
- genes-of-reaction
-
- Description
- Return all genes that encode the enzymes of a given reaction.
- Arguments
-
- rxn
- An instance of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Genes|.
- substrates-of-reaction
-
- Description
- Return all of the reactants and products of a given reaction.
- Arguments
-
- rxn
- An instance of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A list that may consist of children of class
|Compounds|, children of class|Polymer‑Segments|, or strings.
- enzymes-of-reaction
-
- Description
- Return the enzymes that catalyze a given reaction.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - species
- Keyword, Optional A list of species,
such that in a multi-organism PGDB such as MetaCyc, only
proteins found in those organisms will be returned. This list
can include valid
org‑ids, children of class|Organisms|, and strings. Please see the documentation for the'speciesslot‑unitfor more information. Default value isnil. - experimental-only?
- Keyword, Optional When true, only return enzymes that have a non-computational evidence code associated with it.
- local-only-p
- Keyword, Optional When true, only return enzymes that catalyze the specific form of the reaction, as opposed to enzymes that are known to catalyze a more general form (i.e., class) of the reaction.
- Side-Effects
- None.
- Return Value(s)
- A list of children of class
|Proteins|or class|Protein‑RNA‑Complexes|.
- reaction-reactants-and-products
-
- Description
- Return the reactants and products of a reaction, based on a specified direction. The direction can be specified explicity or by giving a pathway as an argument. It is an error to both specify the pathway and the explicit direction. If neither an explicit direction or a pathway is given as an argument, then the direction is computationally inferred from available evidence within the PGDB.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - direction
- Keyword, Optional Can take on the
following values:
- :L2R
- The reaction direction goes from ‘left to
right’, as described in the
|Reactions|instance. - :R2L
- The reaction direction goes from ‘right to
left’; the opposite of what is described in the
|Reactions|instance.
- pwy
- Keyword, Optional
An instance of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- Returns multiple values. The first value is a list of
reactants as determined by the direction of the reaction,
and the second value is a list of the products as determined
by the direction of the reaction. Both lists have items that
are children of class
|Compounds|, children of class|Polymer‑Segments|, or strings.
- reaction-type
-
- Description
- Returns a keyword describing the type of reaction.
- Arguments
-
- rxn
- An instance of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A Common Lisp symbol from the following set:
- :small-molecule
- All substrates are small molecules, or small-molecule classes.
- :transport
- A substrate is marked with different
compartment annotations in the
'leftand'rightslots. - :protein-small-molecule-reaction
- At least one substrate is a protein and at least one is a small molecule.
- :protein-reaction
- All substrates are proteins.
- :trna-reaction
- At least one substrate is a tRNA.
- :null-reaction
- No substrates or reactants are specified.
- :other
- None of the preceding cases apply.
- rxn-without-sequenced-enzyme-p
-
- Description
- A predicate that tests if a given reaction has genes with no associated sequence information.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - complete?
- Optional If true, the predicate will
return true when there is any associated gene without
a sequence. If
nil, the predicate will return true when all associated genes are without a sequence.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- pathway-hole-p
-
- Description
- A predicate that determines if the current reaction is considered to be a ‘pathway hole’, or without an associated enzyme.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - hole-if-any-gene-without-position?
- Keyword, Optional If true, then genes without specified coordinates for the current organism’s genome are not counted when determining the status of the reaction.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- rxn-present?
-
- Description
- A predicate that determines if there is evidence for the occurrence of the given reaction in the current PGDB.
- Arguments
-
- rxn
- An instance of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- rxn-specific-form-of-rxn-p
-
- Description
- A predicate that is true if the given generic reaction is a generalized form of the given specific reaction.
- Arguments
-
- specific-rxn
- A child of the class
|Reactions|. - generic-rxn
- A child of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- nonspecific-forms-of-rxn
-
- Description
- Return all of the generic forms of the given specific reaction. Not every reaction will necessarily have a generic form.
- Arguments
-
- rxn
- An instance of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A list of children of the class
|Reactions|.
- specific-forms-of-rxn
-
- Description
- Return all of the specific forms of the given generic reaction. Not every reaction will necessarily have a specific form.
- Arguments
-
- rxn
- A child of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Reactions|.
- rxn-in-compartment-p
-
- Description
- A predicate that checks if the given reaction is present in a list of cellular compartments.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - compartments
- A list of cellular compartments, as
defined in the Cellular Components Ontology. See frame
'CCO. - default-ok?
- Keyword, Optional
If true, then we return true if the reaction has no
associated compartment information, or one of its associated
locations is a super-class of one of the members of the
compartmentsargument. - pwy
- Keyword, Optional
If supplied, the search for associated enzymes of the
argument
rxnis limited to the given child of|Pathways|. - loose?
- Keyword, Optional
If true, then the compartments
'CCO‑CYTOPLASMand'CCO‑CYTOSOLare treated as being the same compartment.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- compartment-of-rxn
-
- Description
- Returns the compartment of the reaction for non-transport reactions.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - default
- Optional The default compartment
for reactions without any compartment annotations on
their substrates. The default value is
'CCO‑CYTOSOL.
- Side-Effects
- None.
- Return Value(s)
- A child of the class
'CCO.
- compartments-of-reaction
-
- Description
- Returns the compartments associated with the given reaction.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - sides
- Keyword, Optional
The slots of the reaction to consider. The default value
is
'(LEFT RIGHT). - default-compartment
- The default compartment, as
determined by the function
(default‑compartment), which currently is set to'CCO‑CYTOSOL.
- Side-Effects
- None.
- Return Value(s)
- A list of children of the class
'CCO.
- transported-chemicals
-
- Description
- Return the compounds in a transport reaction that change compartments.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - side
- Keyword, Optional The side of the reaction from which to return the transported compound.
- primary-only?
- Keyword, Optional
If true, then filter out common exchangers (currently
defined as
'(PROTON NACPD-1)+). If true, and the only transported compounds are in this list, then the filter doesn’t apply. - from-compartment
- Keyword, Optional
A compartment (child of class
'CCO). If specified, then only return compounds transported from that compartment. - to-compartment
- Keyword, Optional
A compartment (child of class
'CCO). If specified, then only return compounds transported to that compartment. - show-compartment
- Keyword, Optional
A compartment (child of class
'CCO). If specified, and the compound is modified during transport, then only return the form of the compound as found in this compartment.
- Side-Effects
- None.
- Return Value(s)
- A list of children of class
|Compounds|.
- get-predecessors
-
- Description
- Return a list of all reactions that are direct predecessors (i.e., occurr earlier in the pathway) of the given reaction in the given pathway.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - pwy
- A child of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Reactions|.
- get-successors
-
- Description
- Return a list of all reactions that are direct successors (i.e., occurr later in the pathway) of the given reaction in the given pathway.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - pwy
- A child of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Reactions|.
- rxn-w-isozymes-p
-
- Description
- A predicate that tests if a given reaction has any associated isozymes (distinct proteins or protein classes that catalyze the same reaction).
- Arguments
-
- rxn
- An instance of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
5.1.2 Operations on Pathways
- genes-of-pathway
-
- Description
- Return all genes coding for enzymes in the given pathway.
- Arguments
-
- pwy
- An instance of the class
|Pathways|. - sorted?
- Keyword, Optional If true, the genes are sorted in the order in which the corresponding reaction occurrs in the sequence of the pathway.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Genes|.
- enzymes-of-pathway
-
- Description
- Return all enzymes that are present in the given pathway.
- Arguments
-
- pwy
- An instance of the class
|Pathways|. - species
- Keyword, Optional A list of species,
such that in a multi-organism PGDB such as MetaCyc, only
proteins found in those organisms will be returned. This list
can include valid
org‑ids, children of class|Organisms|, and strings. Please see the documentation for the'speciesslot‑unitfor more information. - experimental-only?
- Keyword, Optional When true, only return enzymes that have a non-computational evidence code associated with it.
- sorted?
- Keyword, Optional If true, the enzymes are sorted in the order in which the corresponding reaction occurrs in the sequence of the pathway.
- Side-Effects
- None.
- Return Value(s)
- A list of children of class
|Proteins|or class|Protein‑RNA‑Complexes|.
- 1compounds-of-pathway
-
- Description
- Return all substrates of all reactions that are within the given pathway.
- Arguments
-
- pwy
- An instance of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- A list of children of class
|Compounds|, children of class|Polymer‑Segments|, or strings.
- substrates-of-pathway
-
- Description
- Bearing in mind the direction of all reactions within a pathway, this function returns the substrates of the reactions in four groups: a list of all reactant compounds (compounds occurring on the left side of some reaction in the given pathway), the list of proper reactants (the subset of reactants that are not also products), a list of all products, and a list of all proper products.
- Arguments
-
- pwy
- An instance of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- Four values, each of which is a list of substrates. A
substrate may
be a child of class
|Compounds|, a child of class|Polymer‑Segments|, or a string.
- variants-of-pathway
-
- Description
- Returns all variants of a pathway.
- Arguments
-
- pwy
- An instance of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- A list of instance of the class
|Pathways|.
- pathway-components
-
- Description
- Returns all of the connected components of a pathway. A connected component of a pathway is a set of reactions in the pathway such that for all reactions R1 in the connected component, a predecessor relationship holds between R1 and some other reaction R2 in the connected component, and each connected component is of maximal size. Every pathway will have from 1 to N connected components, where N is the number of reactions in the pathway. Most pathways have one connected component, but not all.
- Arguments
-
- pwy
- An instance of the class
|Pathways|, which is not a super-pathway (i.e., does not have any entries in its'sub‑pathwaysslot). - rxn-list
- Optional
The list of reactions to use as the starting
list of connected component clusters. Defaults to
(get‑slot‑values pwy 'reaction‑list). - pred-list
- Optional
The list of reaction predecessors to
iterate from in order to cluster the reactions in
rxn‑list. Defaults to(get‑slot‑values pwy 'predecessors).
- Side-Effects
- None.
- Return Value(s)
- Returns three values: the connected components as a list of lists of the form
'((r1 r2 r3) (r4 r5) (r6 r7 r8))where each sub-list contains all reactions in one connected component, the number of connected components, and the length of the reaction list.
- noncontiguous-pathway-p
-
- Description
- A predicate that determines if the pathway contains more than one connected component. See function pathway-components for more explanation.
- Arguments
-
- pwy
- An instance of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- rxns-adjacent-in-pwy-p
-
- Description
- A predicate to determine if two given reactions are adjacent to one another in the given pathway.
- Arguments
-
- rxn1
- An instance of the class
|Reactions|. - rxn2
- An instance of the class
|Reactions|. - pwy
- An instance of the class
|Pathways|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
5.1.3 Operations on Enzymatic-Reactions
- cofactors-and-pgroups-of-enzrxn
-
- Description
- Returns the cofactors and prosthetic groups of an enzymatic reaction.
- Arguments
-
- enzrxn
- An instance of the class
|Enzymatic‑Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A list of children of class ’|Chemicals| or strings, representing cofactors and/or prosthetic groups.
- enzrxn-activators
-
- Description
- Returns the list of activators (generally small molecules) of the enzymatic reaction frame.
- Arguments
-
- er
- An instance of the class
|Enzymatic‑Reactions|. - phys-relevant-only?
- Optional
If true, then only return activators that are associated with
|Regulation|instances that have the'physiologically‑relevant?slot set to non-nil.
- Side-Effects
- None.
- Return Value(s)
- A list of children of the class
|Chemicals|.
- enzrxn-inhibitors
-
- Description
- Returns the list of inhibitors (generally small molecules) of the enzymatic reaction frame.
- Arguments
-
- er
- An instance of the class
|Enzymatic‑Reactions|. - phys-relevant-only?
- Optional
If true, then only return inhibitors that are associated with
|Regulation|instances that have the'physiologically‑relevant?slot set to non-nil.
- Side-Effects
- None.
- Return Value(s)
- A list of children of the class
|Chemicals|.
- pathways-of-enzrxn
-
- Description
- Returns the list of pathways in which the given enzymatic reaction participates.
- Arguments
-
- enzrxn
- An instance of the class
|Enzymatic‑Reactions|. - include-super-pwys?
- Keyword, Optional
If non-nil, then not only will the direct pathways in which
enzrxnis associated in be returned, but also any enclosing super-pathways. Ifenzrxnis associated with a reaction that is directly associated with a super-pathway, then the function might return super-pathways even if this option is nil.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Pathways|.
- pathway-allows-enzrxn?
-
- Description
- A predicate which returns a true value if the given pathway allows
the given enzymatic reaction to catalyze the given
reaction. Certain pathways have a list of enzymatic reactions that
are known not to catalyze certain reactions. See the documentation
of slot-unit
'enzyme‑usefor more information. - Arguments
-
- pwy
- An instance of the class
|Pathways|. - rxn
- An instance of the class
|Reactions|. - enzrxn
- An instance of the class
|Enzymatic‑Reactions|. - single-species
- Optional
An instance of the class
|Organisms|If set, thenenzrxnhas the further stricture that it must be an enzymatic reaction present in the organism specified by the value passed tosingle‑species.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
5.1.4 Operations on Proteins
- monomers-of-protein
-
- Description
- Returns the monomers of the given protein complex.
- Arguments
-
- p
- An instance of the class
|Proteins|. - coefficients?
- Keyword, Optional If true, then the second return value of the function will be a list of monomer coefficients. Defaults to true.
- unmodify?
- Keyword, Optional
If true, obtain the monomers of the unmodified form of
p.
- Side-Effects
- None.
- Return Value(s)
- First value is a list of instances of the class ’|Proteins|. If
coefficients?is true, then the second value is the corresponding coefficients of the monomers fromthe first return value.
- base-components-of-protein
-
- Description
- Same as function
#'monomers‑of‑protein, but also returns components of the protein that are RNAs or compounds, not just polypeptides. - Arguments
-
- p
- An instance of the class
|Proteins|. - exclude-small-molecules?
- Keyword, Optional If nil, then small molecule components are also returned. Default value is true.
- Side-Effects
- None.
- Return Value(s)
- Two values. The first value is a list of the components, which can
be instances of the following classes:
|Polypeptides|,|RNA|, and|Compounds|. The second value is a list of the corresponding coefficients of the components in the first value.
- containers-of
-
- Description
- Return all complexes of which the given protein is a direct or indirect component.
- Arguments
-
- protein
- An instance of the class
|Proteins|. - exclude-self?
- Optional
If true, then
proteinwill not be included in the return value.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- protein-or-rna-containers-of
-
- Description
- This function is the same as the function
#'containers‑of, except that it only includes containers that are instances of either class|Protein‑Complexes|, or class|Protein‑RNA‑Complexes|. - Arguments
-
- protein
- An instance of the class
|Proteins|. - exclude-self?
- Optional
If true, then
proteinwill not be included in the return value.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- homomultimeric-containers-of
-
- Description
- This function is the same as the function
#'containers‑of, except that it only includes containers that are homomultimers. - Arguments
-
- protein
- An instance of the class
|Proteins|. - exclude-self?
- Optional
If true, then
proteinwill not be included in the return value.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- polypeptide-or-homomultimer-p
-
- Description
- A predicate that determines if the given protein is a polypeptide or a homomultimer.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- unmodified-form
-
- Description
- Return the unmodified form of the given protein, which might be the same as the given protein.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- An instance of the class
|Proteins|.
- unmodified-or-unbound-form
-
- Description
- Return the unmodified form or unbound (to a small molecule) form of the given protein, which might be the same as the given protein.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- An instance of the class
|Proteins|.
- reduce-modified-proteins
-
- Description
- Given a list of proteins, the function converts all of the proteins to their unmodified form, and then removes any duplicates from the subsequent list.
- Arguments
-
- prots
- A list of instances of the class
|Proteins|. - debind?
- Keyword, Optional When non-nil, the proteins are further simplified by obtaining the unbound form of the protein, if it is bound to a small molecule.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- all-direct-forms-of-protein
-
- Description
- Given a protein, this function will return all of the directly related proteins of its modified and unmodified forms, meaning all of their direct subunits and all of their direct containers.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- all-forms-of-protein
-
- Description
- Given a protein, this function will return all of the
related proteins of its modified and unmodified forms, meaning all
of their subunits and all of their containers. Unlike
#'all‑direct‑forms‑of‑protein, this function is not limited to the direct containers only. - Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- modified-forms
-
- Description
- Returns all modified forms of a protein.
- Arguments
-
- protein
- An instance of the class
|Proteins|. - exclude-self?
- Optional
If true, then
proteinwill not be included in the return value. - all-variants?
- Optional
If true, and
proteinis a modified form, then we return all of the modified forms of the unmodified forms ofprotein.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- modified-and-unmodified-forms
-
- Description
- Returns all of the modified and unmodified forms of the given protein.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- modified-containers
-
- Description
- Returns all containers of a protein (including itself), and all modified forms of the containers.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- top-containers
-
- Description
- Return the top-most containers (i.e., they are not a component of any other protein complex) of the given protein.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- small-molecule-cplxes-of-prot
-
- Description
- Return all of the forms of the given protein that are complexes with small molecules.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- genes-of-protein
-
- Description
- Given a protein, return the set of genes which encode all of the monomers of the protein.
- Arguments
-
- p
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Genes|.
- genes-of-proteins
-
- Description
- The same as
#'genes‑of‑protein, except that it takes a list of proteins and returns a set of genes. - Arguments
-
- p
- A list of instances of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Genes|.
- reactions-of-enzyme
-
- Description
- Return all of the reactions associated with a given protein via enzymatic reactions.
- Arguments
-
- e
- An instance of the class
|Proteins|. - kb
- Keyword, Optional The KB object of the KB in which to find the associated reactions. Defaults to the current PGDB.
- include-specific-forms?
- Keyword, Optional When true, specific forms of associated generic reactions are also returned. Default value is true.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Reactions|.
- species-of-protein
-
- Description
- Get the associated species for the given protein.
- Arguments
-
- p
- A list of instances of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- An instance of the class
|Organisms|, or a string.
- enzyme?
-
- Description
- Predicate that determines whether a specified protein is an enzyme or not.
- Arguments
-
- protein
- An instance of the class
|Proteins|. - type
- Optional
Can take on one of the following values to select more
precisely what is meant by an “enzyme”:
- :any
- Any protein that catalyzes a reaction is considered an enzyme.
- :chemical-change
- If the reactants and products of the catalyzed reactin differ, and not just by their cellular location, then the protein is considered an enzyme.
- :small-molecule
- If the reactants of the catalyzed reaction differ and are small molecules, then the protein is considered an enzyme.
- :transport
- If the protein catalyzes a transport reaction.
- :non-transport
- If the protein only catalyzes non-transport reactions.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- leader-peptide?
-
- Description
- A predicate that determines whether the given protein is a leader peptide.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- protein-p
-
- Description
- A predicate that determines whether the given frame is a protein.
- Arguments
-
- frame
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- complex-p
-
- Description
- A predicate that determines whether the given protein is a protein complex.
- Arguments
-
- frame
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- reactions-of-protein
-
- Description
- Returns all of the associated reactions that the given protein, or its components, catalyzes.
- Arguments
-
- p
- An instance of the class
|Proteins|. - check-protein-components?
- Optional If true, check all components of this protein for catalyzed reactions. Defaults to true.
- check-protein-containers?
- Optional If true, check the containers and modified forms of the protein for catalyzed reactions.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Reactions|.
- protein-in-compartment-p
-
- Description
- A predicate that checks if the given reaction is present in a list of cellular compartments.
- Arguments
-
- rxn
- An instance of the class
|Reactions|. - compartments
- A list of cellular compartments, as
defined in the Cellular Components Ontology. See frame
'CCO. - default-ok?
- Keyword, Optional
If true, then we return true if the reaction has no
associated compartment information, or one of its associated
locations is a super-class of one of the members of the
compartmentsargument. - pwy
- Keyword, Optional
If supplied, the search for associated enzymes of the
argument
rxnis limited to the given child of|Pathways|. - loose?
- Keyword, Optional
If true, then the compartments
'CCO‑CYTOPLASMand'CCO‑CYTOSOLare treated as being the same compartment.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- all-transporters-across
-
- Description
- Returns a list of transport proteins that transport across one of the given membranes.
- Arguments
-
- membranes
- Keyword
Either
:allor a list of instances of the class. Defaults to:all'CCO‑MEMBRANE. - method
- Either
:locationor:reaction‑compartments.:locationwill check the'locationsslot, while:reaction‑compartmentswill examine the compartments of reaction substrates. Default value is:location.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
- autocatalytic-reactions-of-enzyme
-
- Description
- Returns a list of reaction frames, where the protein participates as a substrate of the reaction, and the reaction has no associated Enzymatic Reaction frame. This implies that the protein substrate of the reaction might autocatalyzing the reaction.
- Arguments
-
- prot
- An instance frame of class ’|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Reactions|.
5.1.5 Operations on Genes
- gene-p
-
- Description
- A predicate to determine if the given frame is a gene.
- Arguments
-
- frame
- A frame.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- enzymes-of-gene
-
- Description
- Collects all of the enzymes encoded by the given gene, including modified forms and complexes in which it is a sub-component.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
- all-products-of-gene
-
- Description
- Collects all proteins (not necessarily enzymes) that are encoded by the given gene.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
- reactions-of-gene
-
- Description
- Returns all reactions catalyzed by enzymes encoded by the given gene.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Reactions|.
- pathways-of-gene
-
- Description
- Returns the pathways of enzymes encoded by the given gene.
- Arguments
-
- gene
- An instance of class
|Genes|. - include-super-pwys?
- Keyword, Optional
If non-nil, then not only will the direct pathways in which
geneencodes an enzyme be returned, but also any enclosing super-pathways. Ifgeneis associated with a reaction that is directly associated with a super-pathway, then the function might return super-pathways even if this option is nil.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Pathways|.
- chromosome-of-gene
-
- Description
- Returns the replicon on which the gene is located. If the gene is located on a contig that is, in turn, part of a chromosome, then the contig is returned.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- An instance of class
|Genetic‑Elements|.
- unmodified-gene-product
-
- Description
- Returns the first element of the list returned by the function
unmodified‑gene‑products. This is useful if you are sure that there are no alternative splice forms of your gene. - Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- An instance of either class
|Polypeptides|or'RNA.
- unmodified-gene-products
-
- Description
- Return all of the unmodified gene products (i.e. alternative splice forms) of the given gene.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of either class
|Polypeptides|or'RNA.
- next-gene-on-replicon
-
- Description
- Return the next gene on the replicon.
- Arguments
-
- g
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- Returns two values. The first value is the next gene, or nil if
there is not a next gene (i.e., the gene is at the end of a linear replicon).
The second value is
:lastif the gene is the last gene on a linear replicon.
- previous-gene-on-replicon
-
- Description
- Return the previous gene on the replicon.
- Arguments
-
- g
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- Returns two values. The first value is the previous gene, or nil if
there is not a previous gene (i.e., the gene is at the beginning of a linear replicon).
The second value is
:firstif the gene is the first gene on a linear replicon.
- adjacent-genes?
-
- Description
- Given two genes, this predicate will return true if they are on the same replicon, and adjacent to one another.
- Arguments
-
- g1
- An instance of class
|Genes|. - g2
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- connecting-genes
-
- Description
- Given two genes, if they are on the same replicon, return the replicon, and their numerical ordering among the genes of the replicon, with the smaller number first.
- Arguments
-
- g1
- An instance of class
|Genes|. - g2
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- Three values: the common replicon, the smaller numerical index, and the larger numerical index.
- neighboring-genes-p
-
- Description
- Given two genes, this predicate determines if the two genes are “neighbors”, or within a certain number of genes from one another along the replicon.
- Arguments
-
- g1
- An instance of class
|Genes|. - g2
- An instance of class
|Genes|. - n
- Optional
An integer representing the number of genes
g1andg2can be from one another. Default value is 10.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- gene-clusters
-
- Description
- Groups together genes based on whether each gene is a gene neighbor with other genes.
- Arguments
-
- genes
- A list of instances of class
|Genes|. - max-gap
- Optional
An integer representing the number of genes any pair from
genescan be from one another. Default value is 10.
- Side-Effects
- None.
- Return Value(s)
- A list of lists, where the first element of each sub-list is a gene
from
genes, and the rest of the list are all of the gene neighbors of the first gene.
- rna-coding-gene?
-
- Description
- A predicate that determines if the given gene encodes an RNA.
- Arguments
-
- gene
- An instance of the class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- protein-coding-gene?
-
- Description
- A predicate that determines if the given gene encodes a protein (as opposed to an RNA).
- Arguments
-
- gene
- An instance of the class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- pseudo-gene-p
-
- Description
- A predicate that determines if the given gene is a pseudo-gene.
- Arguments
-
- gene
- An instance of the class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- phantom-gene-p
-
- Description
- A predicate that determines if the given gene is a phantom gene.
- Arguments
-
- gene
- An instance of the class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- dna-binding-site-p
-
- Description
- A predicate that determines if the given frame is an instance of
the class
|DNA‑Binding‑Sites|. - Arguments
-
- gene
- A frame.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- terminatorp
-
- Description
- A predicate that determines if the given object is an instance of
the class
|Terminators|. - Arguments
-
- gene
- A frame.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- operon-of-gene
-
- Description
- Given a gene, return a list of transcription units that form the operon containing the gene.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Transcription‑Units|.
- genes-in-same-operon
-
- Description
- Given a gene, return all other genes in the same operon.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Genes|.
- gene-transcription-units
-
- Description
- Given a gene, return all of the transcription units which contain the gene.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Transcription‑Units|.
- cotranscribed-genes
-
- Description
- Return all co-transcribed genes (i.e., genes which are a part of one or more of the same transcription units) of the given gene.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Genes|.
- terminators-affecting-gene
-
- Description
- Find terminators in the same transcription unit and upstream of the given gene.
- Arguments
-
- gene
- An instance of class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Terminators|.
- chromosome-of-object
-
- Description
- Given an object, the replicon where it is located is returned. If there is no associated replicon for the object, nil is returned. If the object is on more than one replicon, an error is thrown.
- Arguments
-
- object
- An instance of class
|All‑Genes|,|Transcription‑Units|,|Promoters|,|Terminators|,|Misc‑Features|, or|DNA‑Binding‑Sites|.
- Side-Effects
- None.
- Return Value(s)
- An instance of class
|Genetic‑Elements|.
5.1.6 Operations on Regulation Frames
- activation-p
-
- Description
- A predicate that determines if a given regulation frame is describing activation.
- Arguments
-
- reg-frame
- An instance of class
|Regulation|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- inhibition-p
-
- Description
- A predicate that determines if a given regulation frame is describing inhibition.
- Arguments
-
- reg-frame
- An instance of class
|Regulation|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- direct-regulators
-
- Description
- Return all regulators that are connected to a regulated object by a single regulation object.
- Arguments
-
- x
- A frame.
- filter-fn
- Optional A predicate used to filter the regulation objects used to find the regulators.
- Side-Effects
- None.
- Return Value(s)
- A list of frames that regulate
x.
- direct-activators
-
- Description
- Return all activators that are connected to an activated object by a single regulation object.
- Arguments
-
- x
- A frame.
- Side-Effects
- None.
- Return Value(s)
- A list of frames that activate
x.
- direct-inhibitors
-
- Description
- Return all inhibitors that are connected to an inhibited object by a single regulation object.
- Arguments
-
- x
- A frame.
- Side-Effects
- None.
- Return Value(s)
- A list of frames that inhibit
x.
- transcription-factor?
-
- Description
- A predicate that determines if the given protein is a transcription factor, or a component of a transcription factor.
- Arguments
-
- protein
- An instance frame of class
|Proteins|. - include-inactive?
- Keyword, Optional If true, then the function checks to see if any of its components or containers is a transcription factor as well.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- regulator-of-type?
-
- Description
- A predicate that determines if the given protein is a regulator of the specified class.
- Arguments
-
- protein
- An instance frame of class
|Proteins|. - class
- A subclass of
|Regulation|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- regulon-of-protein
-
- Description
- Returns all transcription units regulated by any form of the given protein.
- Arguments
-
- protein
- An instance frame of class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Transcription‑Units|.
- regulation-frame-transcription-units
-
- Description
- Given a regulation object, return the transcription units when one of the regulated entities is a promoter or terminator of the transcription unit.
- Arguments
-
- reg-frame
- An instance of the class
|Regulation‑of‑Transcription|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Transcription‑Units|.
- transcription-unit-regulation-frames
-
- Description
- Returns a list of regulation frames that regulate the transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Regulation|.
- transcription-unit-activation-frames
-
- Description
- Returns a list of regulation frames that activate the transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Regulation|.
- transcription-unit-inhibition-frames
-
- Description
- Returns a list of regulation frames that inhibit the transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Regulation|.
- transcription-units-of-protein
-
- Description
- Return all of the transcription units for which the given protein, or its modified form, acts as a regulator.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Transcription‑Units|.
- genes-regulated-by-protein
-
- Description
- Return all of the genes for which the given protein, or its modified form, acts as a regulator.
- Arguments
-
- protein
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Genes|.
- DNA-binding-sites-of-protein
-
- Description
- Given a transcription factor, return all of its DNA binding sites.
- Arguments
-
- tf
- An instance of the class
|Proteins|. - all-forms?
- Keyword, Optional
When true, then return the DNA binding sites of modified forms
and subunits of
tfas well.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|DNA‑Binding‑Sites|.
- regulator-proteins-of-transcription-unit
-
- Description
- Returns all transcription factors that regulate the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- transcription-factor-ligands
-
- Description
- For a single transcription factor or list of transcription factors, return all transcription factor ligands.
- Arguments
-
- tfs
- An instance or a list of instances of the class
|Proteins|. Iftfsis not the active form, then the active form is determined automatically. - mode
- One of the following values:
:activator,:inhibitor, or:both.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Chemicals|or strings.
- transcription-factor-active-forms
-
- Description
- For a given transcription factor, find all active forms (i.e, form of the protein that regulates) of the transcription factor.
- Arguments
-
- tfs
- An instance of the class
|Proteins|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of the class
|Proteins|.
- genes-regulating-gene
-
- Description
- Return all genes regulating the given gene by means of a transcription factor.
- Arguments
-
- gene
- An instance of the class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Genes|.
- genes-regulated-by-gene
-
- Description
- Return all genes regulated by the given gene by means of a transcription factor.
- Arguments
-
- gene
- An instance of the class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Genes|.
- regulators-of-gene-transcription
-
- Description
- Returns a list of proteins that are regulators of the given gene.
- Arguments
-
- gene
- An instance of the class
|Genes|. - by-function?
- Optional If non-nil, then return two values: a list of activator proteins and a list of inhibitor proteins.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|. Ifby‑function?is non-nil, then two values are returned. The first value is a list of activator proteins, and the second value is a list of inhibitor proteins.
- transcription-unit-activators
-
- Description
- Returns all activator proteins of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
- transcription-unit-inhibitors
-
- Description
- Returns all inhibitor proteins of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
- regulators-of-operon-transcription
-
- Description
- Returns a list of transcription factors of an operon.
- Arguments
-
- operon-list
- A list of instances of the class
|Transcription‑Units|. - by-function?
- Optional If non-nil, then return two values: a list of activator proteins and a list of inhibitor proteins.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|. If the modified form of the protein is the transcription factor, then that is the protein returned.
- transcription-unit-promoter
-
- Description
- Returns the promoter of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- An instance of class
|Promoters|.
- transcription-unit-genes
-
- Description
- Returns the genes of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Genes|.
- transcription-unit-first-gene
-
- Description
- Returns the first gene of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- An instance of class
|Genes|.
- transcription-unit-binding-sites
-
- Description
- Returns the binding sites of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|DNA‑Binding‑Sites|.
- transcription-unit-transcription-factors
-
- Description
- Returns the binding sites of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|DNA‑Binding‑Sites|.
- transcription-unit-mrna-binding-sites
-
- Description
- Returns the mRNA binding sites of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|mRNA‑Binding‑Sites|.
- chromosome-of-operon
-
- Description
- Returns the replicon of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- An instance of class
|Genetic‑Elements|.
- binding-sites-affecting-gene
-
- Description
- Returns all binding sites which are present in the same transcription units as the given gene.
- Arguments
-
- gene
- An instance of the class
|Genes|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|DNA‑Binding‑Sites|.
- binding-site->regulators
-
- Description
- Returns all of the transcription factors of the given binding site.
- Arguments
-
- bsite
- An instance of class
|DNA‑Binding‑Sites|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
- transcription-units-of-promoter
-
- Description
- Returns all transcription units of a given promoter.
- Arguments
-
- promoter
- An instance of class
|Promoters|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Transcription‑Units|.
- promoter-binding-sites
-
- Description
- Returns all of the binding sites associated with the given promoter, across multiple transcription units.
- Arguments
-
- promoter
- An instance of class
|Promoters|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|DNA‑Binding‑Sites|.
- transcription-unit-terminators
-
- Description
- Returns the terminators of the given transcription unit.
- Arguments
-
- operon
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Terminators|.
- containing-tus
-
- Description
- Given a site (whether a DNA binding site, a promoter, a gene, or a terminator) along a transcription unit, return the correspodning transcription units.
- Arguments
-
- site
- An instance of class
|Transcription‑Units|,|mRNA‑Binding‑Sites|,|DNA‑Binding‑Sites|,|Promoters|,|Genes|, or|Terminators|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Transcription‑Units|.
- containing-chromosome
-
- Description
- Given a site (whether a DNA binding site, a promoter, a gene, or a terminator) along a transcription unit, return the correspodning regulon.
- Arguments
-
- site
- An instance of class
|Transcription‑Units|,|mRNA‑Binding‑Sites|,|DNA‑Binding‑Sites|,|Promoters|,|Genes|, or|Terminators|.
- Side-Effects
- None.
- Return Value(s)
- An instance of class
|Genetic‑Elements|.
- binding-site-promoters
-
- Description
- Returns the promoters of the given DNA binding site.
- Arguments
-
- tu
- An instance of the class
|DNA‑Binding‑Sites|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Promoters|.
- transcription-unit-all-components
-
- Description
- Returns all components (binding sites, promoters, genes, terminators) of the given transcription unit.
- Arguments
-
- tu
- An instance of the class
|Transcription‑Units|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Transcription‑Units|,|mRNA‑Binding‑Sites|,|DNA‑Binding‑Sites|,|Promoters|,|Genes|, or|Terminators|.
- binding-site-transcription-units
-
- Description
- Returns all transcription units of a given binding site.
- Arguments
-
- promoter
- An instance of class
|DNA‑Binding‑Sites|or|mRNA‑Binding‑Sites|.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Transcription‑Units|.
5.1.7 Operations on Compounds
- reactions-of-compound
-
- Description
- Return all reactions in which the given compound participates as a substrate.
- Arguments
-
- cpd
- A child of class
|Compounds|. - non-specific-too?
- Keyword, Optional
If non-nil, returns all generic reactions where
cpd, or a parent ofcpd, appears as a substrate. - transport-only?
- Keyword, Optional If non-nil, return only transport reactions.
- compartment
- Keyword, Optional If non-nil, return only reactions within the specified compartment.
- enzymatic?
- Keyword, Optional If non-nil, return only enzymatic reactions.
- Side-Effects
- None.
- Return Value(s)
- A list of children of class
|Reactions|.
- substrate-of-generic-rxn?
-
- Description
- A predicate that determines if a parent of the given compound is a substrate of the given generic reaction.
- Arguments
-
- cpd
- An instance of class
|Compounds|. - rxn
- An instance of class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A boolean value.
- pathways-of-compound
-
- Description
- Returns all pathways in which the given compound appears as a substrate.
- Arguments
-
- cpd
- An instance of class
|Compounds|. - non-specific-too?
- Keyword, Optional
If non-nil, returns all generic reactions where
cpd, or a parent ofcpd, appears as a substrate. - modulators?
- Keyword, Optional
If non-nil, returns pathways where
cpdappears as a regulator as well. - phys-relevant?
- Keyword, Optional
If true, then only return inhibitors that are associated with
|Regulation|instances that have the'physiologically‑relevant?slot set to non-nil. - include-rxns?
- Keyword, Optional If non-nil, then return a list of reaction-pathway pairs.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Pathways|. Ifinclude‑rxns?is non-nil, then a list of lists, where each sub-list consists of an instance of class|Reactions|and an instance of class|Pathways|.
- activated-or-inhibited-by-compound
-
- Description
- Returns all pathways in which the given compound appears as a substrate.
- Arguments
-
- cpds
- An instance or list of instances of class
|Compounds|. - mode
- Keyword, Optional
Represents the type of regulation. Can take on the values of
“+”, “-”, or
'nil. - mechanisms
- Keyword, Optional
Keywords from the
'mechanismslot of the corresponding sub-class of the class|Regulation|. If non-nil, only regulation objects with mechanisms in this list will be explored for regulated objects. - phys-relevant?
- Keyword, Optional
If true, then only return inhibitors that are associated with
|Regulation|instances that have the'physiologically‑relevant?slot set to non-nil. - slots
- Keyword A list of enzymatic reaction slots to
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Enzymatic‑Reactions|.
- tfs-bound-to-compound
-
- Description
- Returns a list of protein complexes that, when bound to the given compound, act as a transcription factor.
- Arguments
-
- cpd
- An instance of class
|Compounds|. - include-inactive?
- Keyword, Optional
If non-nil, then the inactive form of the protein is also
checked. See the function
#'transcription‑factor?for more information.
- Side-Effects
- None.
- Return Value(s)
- A list of instances of class
|Proteins|.
5.1.8 Object Name Manipulation Operations
- get-name-string
-
- Description
- Given an object, compute the string name. The method used to compute the name varies per the object class.
- Arguments
-
- item
- A frame.
- rxn-eqn-as-name?
- Keyword, Optional If non-nil, then we use the reaction equation in string form as the name of the reaction. Defaults to true.
- rxn-common-name-as-name?
- Keyword, Optional If non-nil, then we use the reaction’s common name as the name of the reaction.
- direction
- Keyword, Optional
An argument of
:l2ror:r2lcan be given to specify the desired reaction orientiation when printed in reaction equation form. If this is not provided, then the reaction direction will be determined using pathway evidence. - name-slot
- Keyword, Optional The specified slotunit frame name, as a symbol, will be used for extracting the name of the frame.
- strip-html?
- Keyword, Optional Remove any HTML mark-up from the string form of the object name.
- include-species-strain-name?
- Keyword, Optional Provide proper italicization for the organism strain name.
- italicize-species?
- Keyword, Optional Provide proper italicization for the organism species name.
- short-name?
- Keyword, Optional
If the
'ABBREV‑NAMEslot is populated for the frame, then its value will be used. - species-initials
- Keyword, Optional Print the name of the organism as initials.
- primary-class
- Keyword, Optional Specify explicitly the primary class of the given frame. This can be used to override the internal reasoning of this function, and you can give a suggestion to the function to treat the frame as another class.
- Side-Effects
- None.
- Return Value(s)
- A string representing the name of the frame.
- full-enzyme-name
-
- Description
- Compute the full name of an enzyme as the concatenation of the common name of the protein followed by the common names of its enzymatic reactions. Note that two enzrxns for the same enzyme could have the same common name, so we avoid including the same name twice.
- Arguments
-
- enzyme
- A instance of the class
|Proteins|. - use-frame-name?
- Optional If non-nil, then the frame ID of the enzyme instance is used in computing the enzyme name. Defaults to true.
- name
- Optional A string that bypasses the function, and will be returned as the value of the function.
- activity-names
- Optional
A provided list of strings, that represent
the names of the known catalytic activities of
enzyme.
- Side-Effects
- None.
- Return Value(s)
- A string.
- enzyme-activity-name
-
- Description
- Computes the name of an enzyme in the context of a particular reaction. If the reaction is not provided, then we return the full enzyme name.
- Arguments
-
- enzyme
- An instance of the class
|Proteins|. - reaction
- Optional
An instance of the class
|Reactions|.
- Side-Effects
- None.
- Return Value(s)
- A string.
- subunit-name
-
- Description
-
- Arguments
- Side-Effects
- [
- Return Value(s)
- c
apitalized-gene-name]
- rxn-name-string
- transcription-unit-name
- org-name
- format-organism-name
- frame-organism-name
- orgkb-organism-name
- orgkb-name-equalp
- orgkb-species-name-w-strain
- abbreviate-species-name
- species-initials
6 Code Examples
In this section, we provide a few examples of how to write code in Commonl Lisp that makes use of the Lisp API. This file contains the following sections:
Example Table-Generating Fuctions – Describes utilities that are useful for displaying query results.
Example Queries – Lists a number of example queries
6.1 Example Table-Generating Functions
The functions in this first section are useful for displaying query results. Typically (but not necessarily) a query returns a list of object identifiers. This functions print a more meaningful description of each object, one per line.
;; Given a list of frame names, prints a table of frame names and human-readable ;; names. The optional Format argument selects among different ways of formatting ;; the two columns: :Columns prints fixed-width columns, :Tab separates the ;; columns with a single tab, and :Space separates the columns with a single space. (defun object-table (object-list &optional (format :columns)) (loop for x in object-list for fname = (get-frame-name x) do (ecase format (:columns (format t "~35A ~A~%" fname (get-name-string x))) (:tab (format t "~A ~A~%" fname (get-name-string x))) (:space (format t "~A ~A~%" fname (get-name-string x))) ) ) )
;; Print a list of genes in table form containing, on each line: ;; ecocyc-gene-id ecocyc-gene-name b-number product-name ;; ;; Example usage: (gene-table (find-gene-by-substring "trp")) (defun gene-table (gene-list) (loop for g in gene-list for fname = (get-frame-name g) for prod = (get-slot-value g 'product) for bn = (blattner-gene-id g) do (format t "~20A ~20A ~20A ~A~%" fname (get-slot-value g 'common-name) (or bn "") (if prod (get-name-string prod) "") ) ) )
;; This macro sends all output generated by Body that would normally go ;; to the terminal to the file named by Filename. ;; ;; Example usage: ;; (tofile "~/genes.dat" (gene-table (find-gene-by-substring "trp"))) (defmacro tofile (filename &body body) `(with-open-file (file ,filename :direction :output :if-exists :supersede) (let ((*standard-output* file)) ,@body) ) )
6.2 Example Queries
To run these sample queries, invoke the Pathway Tools from Unix as:
pathway-tools -lisp
The next prompt obtained will be the Lisp listener, to which you can type Lisp expression such as:
(load "examples.lisp")
which will load all of the functions in this file. You can then run a given query by running a function by typing, for example,
(atp-inhibits)
Each example query returns a list of object identifiers. The results are typically printed using a function such as object-table, or else are viewed using the Answer List within the Pathway/Genome Navigator. For example:
(replace-answer-list (oxidoreductases)) (eco)
[click on Next-Answer]
;; Find all oxidoreductases ;; The query iterates through all instances of the EcoCyc class for ;; the Enzyme Commission class 1, which is oxidoreductases, and uses a ;; built-in function that returns all enzymes that catalyze each of those ;; reactions, and appends together the results. (defun oxidoreductases () (loop for x in (get-class-all-instances 'EC-1) append (enzymes-of-reaction x) ) )
;; Find all enzymes for which pyruvate is a substrate ;; The query iterates through all instances of the class Reactions; ;; for those instances whose substrates slot contains pyruvate, ;; the query appends together the enzymes that catalyzes those reactions. (defun pyrsubs () (loop for x in (get-class-all-instances '|Reactions|) when (member-slot-value-p x 'substrates 'pyruvate) append (enzymes-of-reaction x) ) )
;; Find all enzymes for which ATP is an inhibitor (defun atp-inhibits () (loop for x in (get-class-all-instances '|Regulation-of-Enzyme-Activity|) when (and (member-slot-value-p x 'regulator 'atp) (member-slot-value-p x 'mode "-" :test #'fequal) ) collect (get-slot-value (get-slot-value x 'regulated-entity) 'enzyme) ) )
;; Find all enzymes for which pyridoxal phosphate is a prosthetic group. (defun pp-prosthetic () (loop for x in (get-class-all-instances '|Enzymatic-Reactions|) when (member-slot-value-p x 'prosthetic-groups 'Pyridoxal_Phosphate) collect (get-slot-value x 'enzyme) ) )
;; Find all enzymes in the TCA cycle (defun tca-enzymes () (loop for x in (get-slot-values 'TCA 'reaction-list) append (enzymes-of-reaction x) ) )
;; Find all transporters for L-lysine (defun lysine-transporters () (loop for x in (all-rxns :transport) when (member-slot-value-p x 'substrates 'Lys) append (enzymes-of-reaction x) ) )
;; Find all enzymes that accept glutamine and ammonia as ;; alternative substrates (defvar *enzymes*) (defun gltamm-subs () (setq *enzymes* nil) (loop for x in (get-class-all-instances '|Enzymatic-Reactions|) do (loop for item in (get-slot-values x 'alternative-substrates) when (or (and (fequal 'AMMONIA (first item)) (find 'GLN (rest item)) ) (and (fequal 'GLN (first item)) (find 'AMMONIA (rest item)) ) ) do (push (get-slot-value x 'enzyme) *enzymes*) ) ) )
;; Find all genes that contain a given name as their common name or ;; as a synonym: (defun find-gene-by-name (name) (loop for g in (get-class-all-instances '|Genes|) when (member-slot-value-p g 'names name) collect g) )
;; Find all that genes that contain a given substring within their ;; common name or synonym list. The use of the string-equal test ;; ensures case-insensitve matching. The result variable is used ;; because if we use a collect within the second loop, its result ;; will not be collected by the first loop. (defun find-gene-by-substring (substring) (let (result) (loop for g in (get-class-all-instances '|Genes|) do (loop for name in (get-slot-values g 'names) when (search substring name :test #'string-equal) do (pushnew g result) ) ) result ) )
;; Find genes located between 20 and 30 centisomes on the map (loop for gene in (get-class-all-instances '|Genes|) for pos = (get-slot-value gene 'centisome-position) when (and pos (> pos 20) (< pos 30)) collect gene)
;; The preceding query returns a list of genes. To run the Pathway/ ;; Genome Navigator with those genes on the Answer list, ;; evaluate the following. The "*" means "the result returned by ;; the last expression evaluated". (eco :answer-list *)
;; Find reactions involving pyruvate as a substrate (loop for rxn in (get-class-all-instances '|Reactions|) when (member 'pyruvate (get-slot-values rxn 'substrates)) collect rxn)
;; Find all genes whose products catalyze a reaction involving ;; pyruvate as a substrate (loop for rxn in (get-class-all-instances '|Reactions|) for genes = (genes-of-reaction rxn) when (member-slot-value-p rxn 'substrates 'pyruvate) append genes)
;; Find all enzymes that use pyridoxal phosphate as a cofactor ;; or prosthetic group (loop for protein in (get-class-all-instances '|Proteins|) for enzrxn = (get-slot-value protein 'enzymatic-reaction) when (and enzrxn (or (member-slot-value-p enzrxn 'cofactors 'pyridoxal_phosphate) (member-slot-value-p enzrxn 'prosthetic-groups 'pyridoxal_phosphate) )) collect protein)